Update, 2015-11-30 It appears that NOAA has gone through and upgraded all of the OISST files to the newer version of the NetCDF file format. As a result, the functions outlined in this post don’t work any longer. Instead, see the updated functions in my newer post, https://lukemiller.org/index.php/2014/11/extracting-noaa-sea-surface-temperatures-with-ncdf4/. The concepts are the same as described here, but the newer functions use the ncdf4 package to access the newer NetCDF file format.
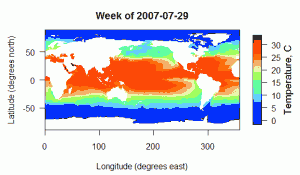
NOAA’s Physical Sciences Division produces a global map of weekly averaged sea surface temperatures, derived from satellite sources. The data can be found at http://www.esrl.noaa.gov/psd/data/gridded/data.noaa.oisst.v2.html. I’ve put together a R script that will extract data from a region of interest and optionally plot it for you. The input data file is a 135+ MB netCDF file found at the link above.
The OISSTv2 data are arranged in a 1° x 1° grid covering the entire world, and a weekly average has been calculated for each grid cell for each 7-day period starting Jan 1, 1990, in the file sst.wkmean.1990-present.nc. You will also want to grab the land-sea mask file lsmask.nc as well. The land-sea mask is necessary to remove the spurious temperature values that fall on the continents. In the script below I use the land-sea mask to simply convert all points over land into NA values in the output matrix.
This script makes use of two add-on packages available from the CRAN repositories. First, the ncdf package, written by David Pierce and maintained by Brian Ripley, is used to open the netCDF file and extract the desired data. The fields package, written by Reinhard Furrer, Douglas Nychka and Stephen Sain, provides the tools to plot the data with a color scale bar alongside the plot.
The NOAA_OISST_extraction.R script can be downloaded here. Please see the newer functions described in https://lukemiller.org/index.php/2014/11/extracting-noaa-sea-surface-temperatures-with-ncdf4/
# Filename: NOAA_OISST_extraction.R # # Author: Luke Miller Mar 1, 2011 ############################################################################### library(ncdf) # For info on the NOAA Optimum Interpolated Sea Surface Temperature V2 (OISST): # http://www.esrl.noaa.gov/psd/data/gridded/data.noaa.oisst.v2.html # This script works on the OISST sea surface temperature netCDF file called # sst.wkmean.1990-present.nc, which must be downloaded into the current # working directory (file size is >135MB currently) # The OISST grid layout is 1 degree latitudes from 89.5 to -89.5 degrees north. # The longitude grid is 1 degree bins, 0.5 to 359.5 degrees east. # The SST values are given in degrees Celsius. # Time values are "days since 1800-1-1 00:00", starting from 69395 # Time delta_t is 0000-00-07 00:00, indices range from 0 to 1103 currently # The time values in the sst field give the 1st day of the week for each # weekly temperature value (though the data are meant to be centered on # Wednesday of that week), starting at 69395 = 12/31/1989. # Use as.Date(69395,origin = '1800-1-1') to convert the netCDF day value to a # human readable form. lats = seq(89.5,-89.5,-1) #generate set of grid cell latitudes (center of cell) lons = seq(0.5,359.5,1) #generate set of grid cell longitudes (center of cell) #Ask user to enter the boundaries of the search area cat("Enter longitude of western edge of search area (degrees east 0 to 359)\n") lon1 = scan(what = numeric(0),n = 1) cat("Enter longitude of eastern edge of search area (degrees east 0 to 359)\n") lon2 = scan(what = numeric(0),n = 1) cat("Enter latitude of northern edge\n") cat("of search area (degrees north, 89.5 to -89.5\n") lat1 = scan(what = numeric(0),n = 1) cat("Enter latitude of southern edge\n") cat("of search area (degrees north, 89.5 to -89.5\n") lat2 = scan(what = numeric(0),n = 1) lon1a = which.min(abs(lon1 - lons)) #get index of nearest longitude value lon2a = which.min(abs(lon2 - lons)) #get index of nearest longitude value lat1a = which.min(abs(lat1 - lats)) #get index of nearest latitude value lat2a = which.min(abs(lat2 - lats)) #get index of nearest latitude value #The lon/lat 1a/2a values should now correspond to indices in the netCDF file #for the desired grid cell. nlon = (lon2a - lon1a) + 1 #get number of longitudes to extract nlat = (lat2a - lat1a) + 1 #get number of latitudes to extract #Ask the user to enter the dates of interest cat("Enter the starting date in the form: 1990-1-31\n") date1 = scan(what = character(0),n = 1) cat("Enter the ending date in the form: 1990-1-31\n") date2 = scan(what = character(0),n = 1) #date1 = '2007-08-01' #date2 = '2007-09-01' date1 = as.Date(date1, format = "%Y-%m-%d") #convert to Date object date2 = as.Date(date2, format = "%Y-%m-%d") #convert to Date object #Open the netCDF file for reading. nc = open.ncdf("sst.wkmean.1990-present.nc") #print.ncdf(nc) will show info about the structure of the netCDF file #Extract available dates from netCDF file ncdates = nc$dim$time$vals ncdates = as.Date(ncdates,origin = '1800-1-1') #available time points in nc date1a = which.min(abs(date1 - ncdates)) #get index of nearest time point date2a = which.min(abs(date2 - ncdates)) #get index of nearest time point ndates = (date2a - date1a) + 1#get number of time steps to extract #Extract the data from the netCDF file to a matrix or array called 'sstout'. sstout = get.var.ncdf(nc, varid = 'sst', start = c(lon1a,lat1a,date1a), count = c(nlon,nlat,ndates)) #If you only retrieve one time point, sstout will be a 2D matrix with #longitudes running down the rows, and latitudes across the columns. #For example, Column 1, sstout[1:nrow(sstout),1], will contain sst values for #each of the longitude values at the northern-most latitude in your search #area, with the first row, sstout[1,1], being the western-most longitude, and #the last row being the eastern edge of your search area. #If you retrieve multiple time points, sstout will be a 3D array, where time is #the 3rd dimension. Lat/lon will be arranged the same as the 2D case. #The vector 'datesout' will hold the Date values associated with each set of #sst values in the sstout array, should you need to access them. datesout = ncdates[date1a:date2a] ############################################################################### ############################################################################### # The NOAA OISST files contain sea surface temperatures for the entire globe, # including on the continents. This clearly isn't right, so they also supply a # land-sea mask file in netCDF format at the website listed at the start of # this script. # We use the values (0 or 1) stored in the mask file to turn all of the # continent areas into NA's. nc2 = open.ncdf('lsmask.nc') #open land-sea mask mask = get.var.ncdf(nc2, varid = "mask",start = c(lon1a,lat1a,1), count = c(nlon,nlat,1)) #get land-sea mask values (0 or 1) mask = ifelse(mask == 0,NA,1) #replace 0's with NA's if (is.matrix(sstout)) { #if there is only 1 time point (2D matrix) sstout = sstout * mask #all masked values become NA } if (!is.matrix(sstout)) { #if ssout is a 3D matrix dims = dim(sstout) for (i in 1:dims[3]){ sstout[,,i] = sstout[,,i] * mask #all masked values become NA } } ############################################################################### ############################################################################### #Now we can produce a plot of the data. library(fields) goodlons = lons[lon1a:lon2a] goodlats = lats[lat1a:lat2a] #plot the 1st time point if (is.matrix(sstout)){ #if there is only one time point par(mar = c(5,5,4,6)) #widen margins image(goodlons,rev(goodlats),sstout[,], ylim = rev(range(goodlats)), #reverse y-axis col = tim.colors(32), yaxt = "n", xaxt = "n", ylab = "Latitude (degrees north)", xlab = "Longitude (degrees east)", main = paste("Week of ",datesout[1],sep = "")) axis(1,at = pretty(goodlons),labels = pretty(goodlons), las = 1) axis(2,at = rev(pretty(goodlats)),labels = pretty(goodlats), las = 1) #image.plot from the fields package inserts a color bar image.plot(zlim = range(sstout[,],na.rm = TRUE),nlevel = 32, legend.only = TRUE, horizontal = FALSE, col = tim.colors(32), legend.args = list(text = "Temperature, C", cex = 1.2, side = 4, line = 2)) } else { #If there is more than one time point, just plot the first time point par(mar = c(4.5,4.5,3,6)) #widen margins image(goodlons,rev(goodlats),sstout[,,1], ylim = rev(range(goodlats)), #reverse y-axis col = tim.colors(32), yaxt = "n", xaxt = "n", ylab = "Latitude (degrees north)", xlab = "Longitude (degrees east)", main = paste("Week of ",datesout[1],sep = "")) axis(1,at = pretty(goodlons),labels = pretty(goodlons), las = 1) axis(2,at = rev(pretty(goodlats)),labels = pretty(goodlats), las = 1) #image.plot from the fields package inserts a color bar image.plot(zlim = range(sstout[,,1],na.rm = TRUE),nlevel = 32, legend.only = TRUE, horizontal = FALSE, col = tim.colors(32), legend.args = list(text = "Temperature, C", cex = 1.2, side = 4, line = 2)) }